-
Jun 19, 2023
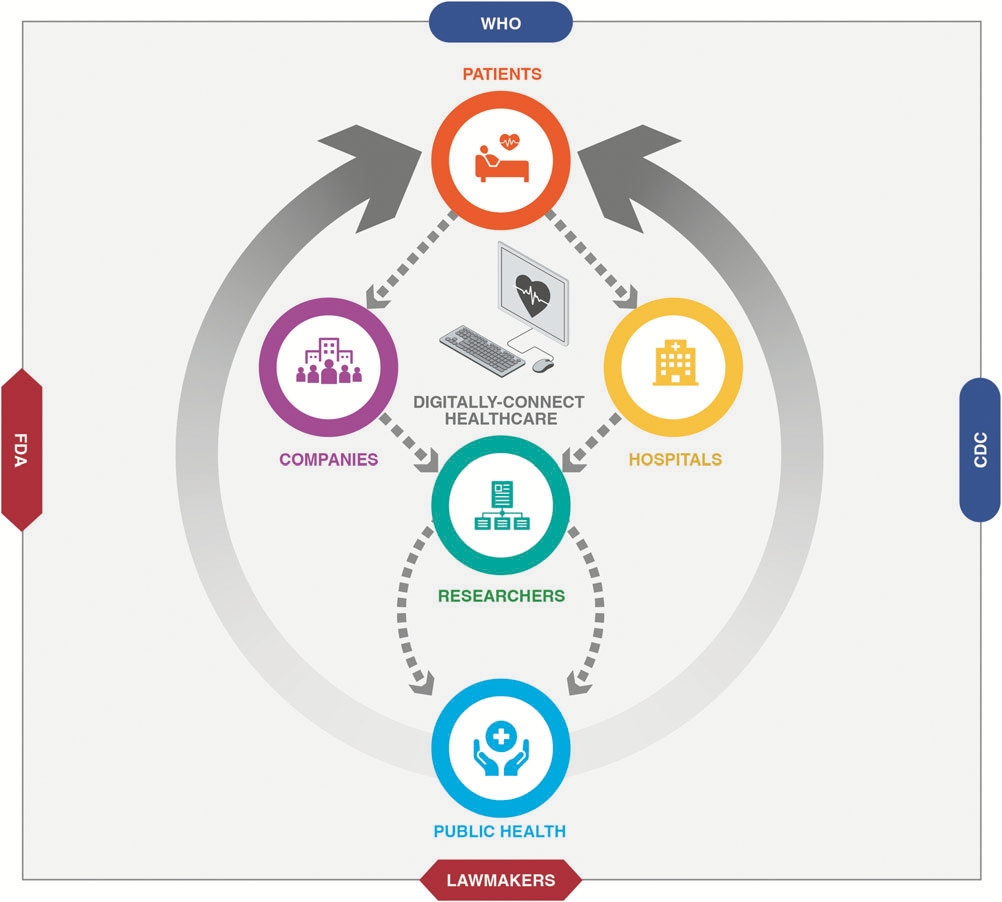 The COVID-19 pandemic has challenged both health care administrators and public health policymakers in unprecedented ways. The dearth of actionable clinical data nationwide impeded prompt evidence-based policymaking and adjustments to health care systems. This highlighted the importance of accessible high-quality real-time clinical data at the local and national level for tackling emerging public health threats. Current methods of data collection for public health are tedious and time-consuming, limiting the speed at which effective measures are determined. Meanwhile, diagnostic companies are increasingly embracing digital technologies, which could put anonymized aggregate diagnostic data, and effective analytics at the heart of health care policy and management. The key to achieving that is—in part—crafting effective public–private partnerships at the federal and international level. Carefully carving out the terms of such partnerships will be crucial to their success. The benefits for governments and their citizens are likely to be worthwhile as public health threats continue to arise, spending remains overstretched, and health care systems overburdened.
The COVID-19 pandemic has challenged both health care administrators and public health policymakers in unprecedented ways. The dearth of actionable clinical data nationwide impeded prompt evidence-based policymaking and adjustments to health care systems. This highlighted the importance of accessible high-quality real-time clinical data at the local and national level for tackling emerging public health threats. Current methods of data collection for public health are tedious and time-consuming, limiting the speed at which effective measures are determined. Meanwhile, diagnostic companies are increasingly embracing digital technologies, which could put anonymized aggregate diagnostic data, and effective analytics at the heart of health care policy and management. The key to achieving that is—in part—crafting effective public–private partnerships at the federal and international level. Carefully carving out the terms of such partnerships will be crucial to their success. The benefits for governments and their citizens are likely to be worthwhile as public health threats continue to arise, spending remains overstretched, and health care systems overburdened.
-
Mar 17, 2023
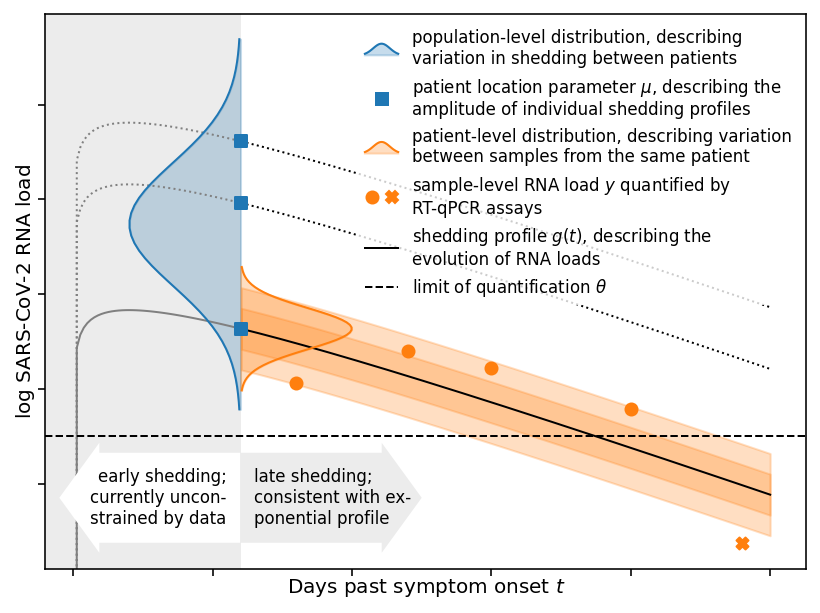 The concentration of SARS-CoV-2 RNA in faeces is not well characterised, posing challenges for quantitative wastewater-based epidemiology (WBE). We developed hierarchical models for faecal RNA shedding and fitted them to data from six studies. A mean concentration of 1.9×10⁶ mL⁻¹ (2.3×10⁵–2.0×10⁸ 95% credible interval) was found among unvaccinated inpatients, not considering differences in shedding between viral variants. Limits of quantification could account for negative samples based on Bayesian model comparison. Inpatients represented the tail of the shedding profile with a half-life of 34 hours (28–43 95% credible interval), suggesting that WBE can be a leading indicator for clinical presentation. Shedding among inpatients could not explain the high RNA concentrations found in wastewater, consistent with more abundant shedding during the early infection course.
The concentration of SARS-CoV-2 RNA in faeces is not well characterised, posing challenges for quantitative wastewater-based epidemiology (WBE). We developed hierarchical models for faecal RNA shedding and fitted them to data from six studies. A mean concentration of 1.9×10⁶ mL⁻¹ (2.3×10⁵–2.0×10⁸ 95% credible interval) was found among unvaccinated inpatients, not considering differences in shedding between viral variants. Limits of quantification could account for negative samples based on Bayesian model comparison. Inpatients represented the tail of the shedding profile with a half-life of 34 hours (28–43 95% credible interval), suggesting that WBE can be a leading indicator for clinical presentation. Shedding among inpatients could not explain the high RNA concentrations found in wastewater, consistent with more abundant shedding during the early infection course.
-
Mar 15, 2023
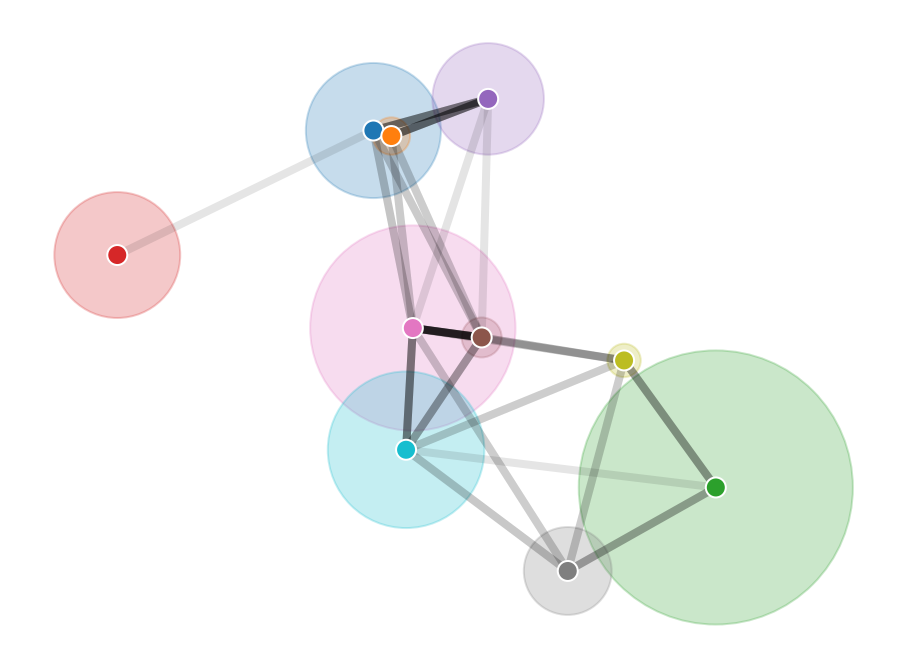 Large-scale network data can pose computational challenges, be expensive to acquire, and compromise the privacy of individuals in social networks. We show that the locations and scales of latent space cluster models can be inferred from the number of connections between groups alone. We demonstrate this modelling approach using synthetic data and apply it to friendships between students collected as part of the Add Health study, eliminating the need for node-level connection data. The method thus protects the privacy of individuals and simplifies data sharing. It also offers performance advantages over node-level latent space models because the computational cost scales with the number of clusters rather than the number of nodes.
Large-scale network data can pose computational challenges, be expensive to acquire, and compromise the privacy of individuals in social networks. We show that the locations and scales of latent space cluster models can be inferred from the number of connections between groups alone. We demonstrate this modelling approach using synthetic data and apply it to friendships between students collected as part of the Add Health study, eliminating the need for node-level connection data. The method thus protects the privacy of individuals and simplifies data sharing. It also offers performance advantages over node-level latent space models because the computational cost scales with the number of clusters rather than the number of nodes.
-
Jan 24, 2023
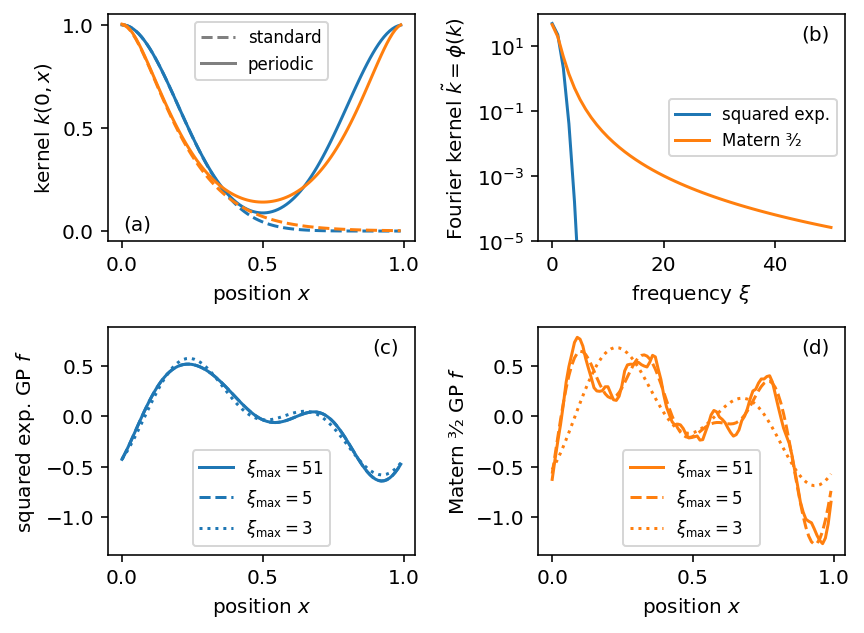 Gaussian processes (GPs) are sophisticated distributions to model functional data. Whilst theoretically appealing, they are computationally cumbersome except for small datasets. We implement two methods for scaling GP inference in Stan: First, a general sparse approximation using a directed acyclic dependency graph. Second, a fast, exact method for regularly spaced data modeled by GPs with stationary kernels using the fast Fourier transform. Based on benchmark experiments, we offer guidance for practitioners to decide between different methods and parameterizations. We consider two real-world examples to illustrate the package. The implementation follows Stan’s design and exposes performant inference through a familiar interface. Full posterior inference for more than ten thousand data points is feasible on a laptop in less than 20 seconds.
Gaussian processes (GPs) are sophisticated distributions to model functional data. Whilst theoretically appealing, they are computationally cumbersome except for small datasets. We implement two methods for scaling GP inference in Stan: First, a general sparse approximation using a directed acyclic dependency graph. Second, a fast, exact method for regularly spaced data modeled by GPs with stationary kernels using the fast Fourier transform. Based on benchmark experiments, we offer guidance for practitioners to decide between different methods and parameterizations. We consider two real-world examples to illustrate the package. The implementation follows Stan’s design and exposes performant inference through a familiar interface. Full posterior inference for more than ten thousand data points is feasible on a laptop in less than 20 seconds.
-
Jan 20, 2023
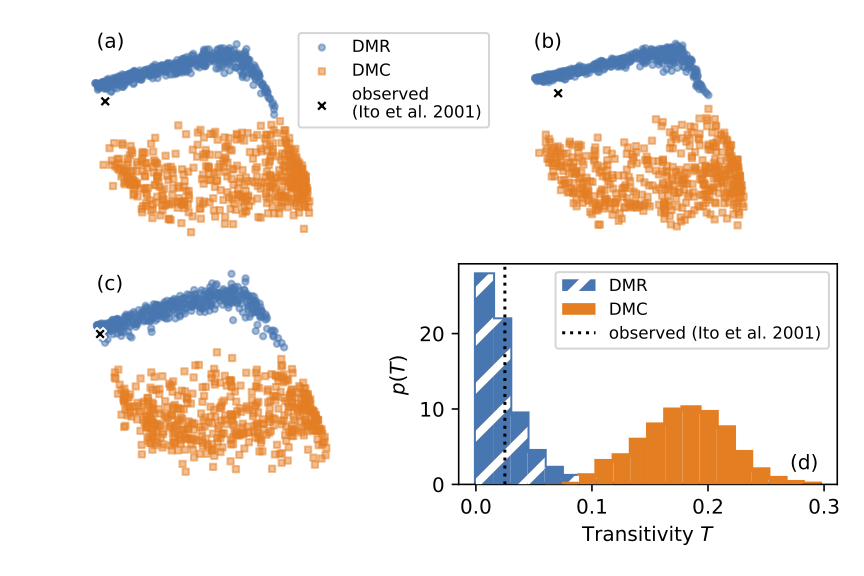 Selecting a small set of informative features from a large number of possibly noisy candidates is a challenging problem with many applications in machine learning and approximate Bayesian computation. In practice, the cost of computing informative features also needs to be considered. This is particularly important for networks because the computational costs of individual features can span several orders of magnitude. We addressed this issue for the network model selection problem using two approaches. First, we adapted nine feature selection methods to account for the cost of features. We show for two classes of network models that the cost can be reduced by two orders of magnitude without considerably affecting classification accuracy (proportion of correctly identified models). Second, we selected features using pilot simulations with smaller networks. This approach reduced the computational cost by a factor of 50 without affecting classification accuracy. To demonstrate the utility of our approach, we applied it to three different yeast protein interaction networks and identified the best-fitting duplication divergence model. Supplementary materials, including computer code to reproduce our results, are available online.
Selecting a small set of informative features from a large number of possibly noisy candidates is a challenging problem with many applications in machine learning and approximate Bayesian computation. In practice, the cost of computing informative features also needs to be considered. This is particularly important for networks because the computational costs of individual features can span several orders of magnitude. We addressed this issue for the network model selection problem using two approaches. First, we adapted nine feature selection methods to account for the cost of features. We show for two classes of network models that the cost can be reduced by two orders of magnitude without considerably affecting classification accuracy (proportion of correctly identified models). Second, we selected features using pilot simulations with smaller networks. This approach reduced the computational cost by a factor of 50 without affecting classification accuracy. To demonstrate the utility of our approach, we applied it to three different yeast protein interaction networks and identified the best-fitting duplication divergence model. Supplementary materials, including computer code to reproduce our results, are available online.
 The COVID-19 pandemic has challenged both health care administrators and public health policymakers in unprecedented ways. The dearth of actionable clinical data nationwide impeded prompt evidence-based policymaking and adjustments to health care systems. This highlighted the importance of accessible high-quality real-time clinical data at the local and national level for tackling emerging public health threats. Current methods of data collection for public health are tedious and time-consuming, limiting the speed at which effective measures are determined. Meanwhile, diagnostic companies are increasingly embracing digital technologies, which could put anonymized aggregate diagnostic data, and effective analytics at the heart of health care policy and management. The key to achieving that is—in part—crafting effective public–private partnerships at the federal and international level. Carefully carving out the terms of such partnerships will be crucial to their success. The benefits for governments and their citizens are likely to be worthwhile as public health threats continue to arise, spending remains overstretched, and health care systems overburdened.
The COVID-19 pandemic has challenged both health care administrators and public health policymakers in unprecedented ways. The dearth of actionable clinical data nationwide impeded prompt evidence-based policymaking and adjustments to health care systems. This highlighted the importance of accessible high-quality real-time clinical data at the local and national level for tackling emerging public health threats. Current methods of data collection for public health are tedious and time-consuming, limiting the speed at which effective measures are determined. Meanwhile, diagnostic companies are increasingly embracing digital technologies, which could put anonymized aggregate diagnostic data, and effective analytics at the heart of health care policy and management. The key to achieving that is—in part—crafting effective public–private partnerships at the federal and international level. Carefully carving out the terms of such partnerships will be crucial to their success. The benefits for governments and their citizens are likely to be worthwhile as public health threats continue to arise, spending remains overstretched, and health care systems overburdened. The concentration of SARS-CoV-2 RNA in faeces is not well characterised, posing challenges for quantitative wastewater-based epidemiology (WBE). We developed hierarchical models for faecal RNA shedding and fitted them to data from six studies. A mean concentration of 1.9×10⁶ mL⁻¹ (2.3×10⁵–2.0×10⁸ 95% credible interval) was found among unvaccinated inpatients, not considering differences in shedding between viral variants. Limits of quantification could account for negative samples based on Bayesian model comparison. Inpatients represented the tail of the shedding profile with a half-life of 34 hours (28–43 95% credible interval), suggesting that WBE can be a leading indicator for clinical presentation. Shedding among inpatients could not explain the high RNA concentrations found in wastewater, consistent with more abundant shedding during the early infection course.
The concentration of SARS-CoV-2 RNA in faeces is not well characterised, posing challenges for quantitative wastewater-based epidemiology (WBE). We developed hierarchical models for faecal RNA shedding and fitted them to data from six studies. A mean concentration of 1.9×10⁶ mL⁻¹ (2.3×10⁵–2.0×10⁸ 95% credible interval) was found among unvaccinated inpatients, not considering differences in shedding between viral variants. Limits of quantification could account for negative samples based on Bayesian model comparison. Inpatients represented the tail of the shedding profile with a half-life of 34 hours (28–43 95% credible interval), suggesting that WBE can be a leading indicator for clinical presentation. Shedding among inpatients could not explain the high RNA concentrations found in wastewater, consistent with more abundant shedding during the early infection course. Large-scale network data can pose computational challenges, be expensive to acquire, and compromise the privacy of individuals in social networks. We show that the locations and scales of latent space cluster models can be inferred from the number of connections between groups alone. We demonstrate this modelling approach using synthetic data and apply it to friendships between students collected as part of the Add Health study, eliminating the need for node-level connection data. The method thus protects the privacy of individuals and simplifies data sharing. It also offers performance advantages over node-level latent space models because the computational cost scales with the number of clusters rather than the number of nodes.
Large-scale network data can pose computational challenges, be expensive to acquire, and compromise the privacy of individuals in social networks. We show that the locations and scales of latent space cluster models can be inferred from the number of connections between groups alone. We demonstrate this modelling approach using synthetic data and apply it to friendships between students collected as part of the Add Health study, eliminating the need for node-level connection data. The method thus protects the privacy of individuals and simplifies data sharing. It also offers performance advantages over node-level latent space models because the computational cost scales with the number of clusters rather than the number of nodes. Gaussian processes (GPs) are sophisticated distributions to model functional data. Whilst theoretically appealing, they are computationally cumbersome except for small datasets. We implement two methods for scaling GP inference in Stan: First, a general sparse approximation using a directed acyclic dependency graph. Second, a fast, exact method for regularly spaced data modeled by GPs with stationary kernels using the fast Fourier transform. Based on benchmark experiments, we offer guidance for practitioners to decide between different methods and parameterizations. We consider two real-world examples to illustrate the package. The implementation follows Stan’s design and exposes performant inference through a familiar interface. Full posterior inference for more than ten thousand data points is feasible on a laptop in less than 20 seconds.
Gaussian processes (GPs) are sophisticated distributions to model functional data. Whilst theoretically appealing, they are computationally cumbersome except for small datasets. We implement two methods for scaling GP inference in Stan: First, a general sparse approximation using a directed acyclic dependency graph. Second, a fast, exact method for regularly spaced data modeled by GPs with stationary kernels using the fast Fourier transform. Based on benchmark experiments, we offer guidance for practitioners to decide between different methods and parameterizations. We consider two real-world examples to illustrate the package. The implementation follows Stan’s design and exposes performant inference through a familiar interface. Full posterior inference for more than ten thousand data points is feasible on a laptop in less than 20 seconds. Selecting a small set of informative features from a large number of possibly noisy candidates is a challenging problem with many applications in machine learning and approximate Bayesian computation. In practice, the cost of computing informative features also needs to be considered. This is particularly important for networks because the computational costs of individual features can span several orders of magnitude. We addressed this issue for the network model selection problem using two approaches. First, we adapted nine feature selection methods to account for the cost of features. We show for two classes of network models that the cost can be reduced by two orders of magnitude without considerably affecting classification accuracy (proportion of correctly identified models). Second, we selected features using pilot simulations with smaller networks. This approach reduced the computational cost by a factor of 50 without affecting classification accuracy. To demonstrate the utility of our approach, we applied it to three different yeast protein interaction networks and identified the best-fitting duplication divergence model. Supplementary materials, including computer code to reproduce our results, are available online.
Selecting a small set of informative features from a large number of possibly noisy candidates is a challenging problem with many applications in machine learning and approximate Bayesian computation. In practice, the cost of computing informative features also needs to be considered. This is particularly important for networks because the computational costs of individual features can span several orders of magnitude. We addressed this issue for the network model selection problem using two approaches. First, we adapted nine feature selection methods to account for the cost of features. We show for two classes of network models that the cost can be reduced by two orders of magnitude without considerably affecting classification accuracy (proportion of correctly identified models). Second, we selected features using pilot simulations with smaller networks. This approach reduced the computational cost by a factor of 50 without affecting classification accuracy. To demonstrate the utility of our approach, we applied it to three different yeast protein interaction networks and identified the best-fitting duplication divergence model. Supplementary materials, including computer code to reproduce our results, are available online.