-
Mar 15, 2023
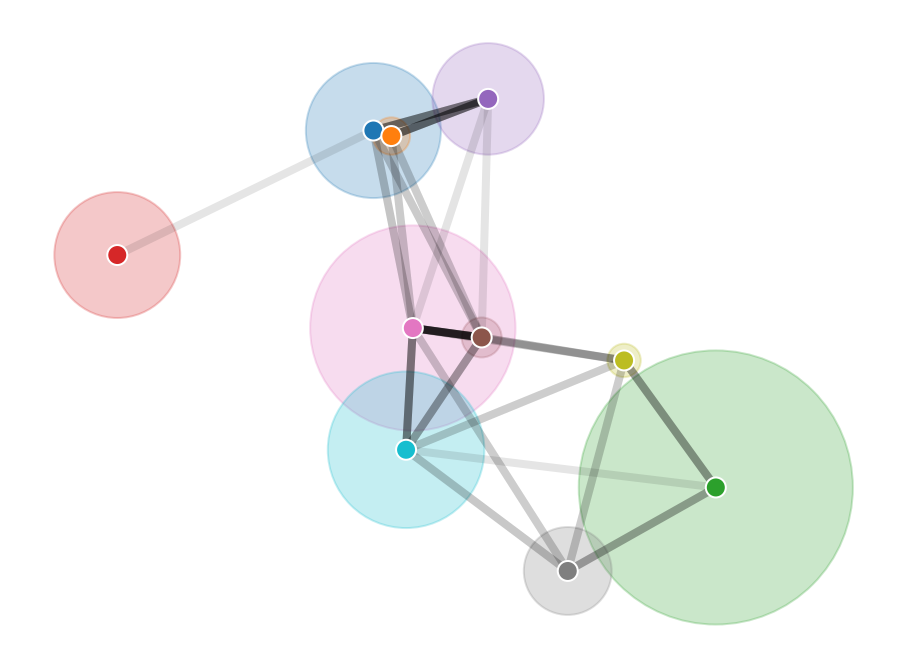 Large-scale network data can pose computational challenges, be expensive to acquire, and compromise the privacy of individuals in social networks. We show that the locations and scales of latent space cluster models can be inferred from the number of connections between groups alone. We demonstrate this modelling approach using synthetic data and apply it to friendships between students collected as part of the Add Health study, eliminating the need for node-level connection data. The method thus protects the privacy of individuals and simplifies data sharing. It also offers performance advantages over node-level latent space models because the computational cost scales with the number of clusters rather than the number of nodes.
Large-scale network data can pose computational challenges, be expensive to acquire, and compromise the privacy of individuals in social networks. We show that the locations and scales of latent space cluster models can be inferred from the number of connections between groups alone. We demonstrate this modelling approach using synthetic data and apply it to friendships between students collected as part of the Add Health study, eliminating the need for node-level connection data. The method thus protects the privacy of individuals and simplifies data sharing. It also offers performance advantages over node-level latent space models because the computational cost scales with the number of clusters rather than the number of nodes.
-
Jan 24, 2023
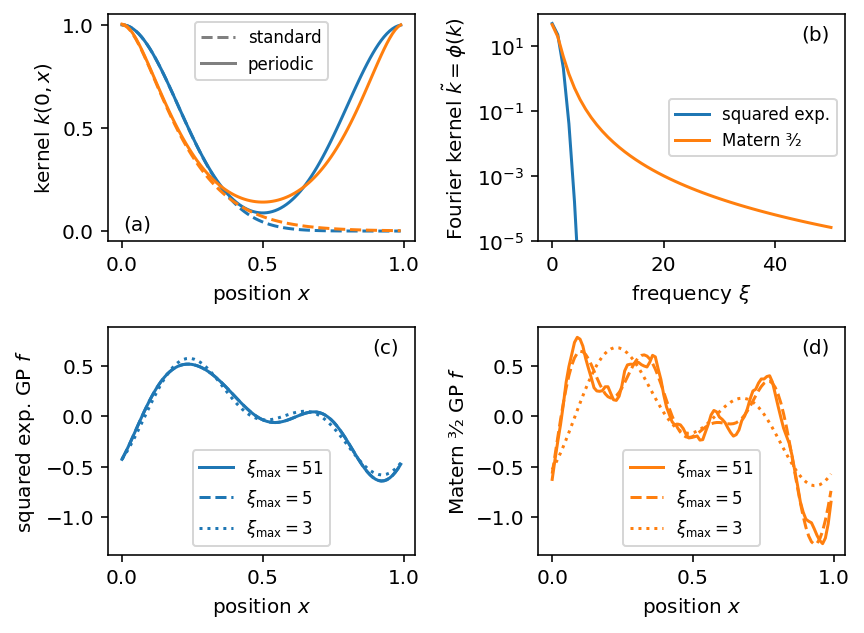 Gaussian processes (GPs) are sophisticated distributions to model functional data. Whilst theoretically appealing, they are computationally cumbersome except for small datasets. We implement two methods for scaling GP inference in Stan: First, a general sparse approximation using a directed acyclic dependency graph. Second, a fast, exact method for regularly spaced data modeled by GPs with stationary kernels using the fast Fourier transform. Based on benchmark experiments, we offer guidance for practitioners to decide between different methods and parameterizations. We consider two real-world examples to illustrate the package. The implementation follows Stan’s design and exposes performant inference through a familiar interface. Full posterior inference for more than ten thousand data points is feasible on a laptop in less than 20 seconds.
Gaussian processes (GPs) are sophisticated distributions to model functional data. Whilst theoretically appealing, they are computationally cumbersome except for small datasets. We implement two methods for scaling GP inference in Stan: First, a general sparse approximation using a directed acyclic dependency graph. Second, a fast, exact method for regularly spaced data modeled by GPs with stationary kernels using the fast Fourier transform. Based on benchmark experiments, we offer guidance for practitioners to decide between different methods and parameterizations. We consider two real-world examples to illustrate the package. The implementation follows Stan’s design and exposes performant inference through a familiar interface. Full posterior inference for more than ten thousand data points is feasible on a laptop in less than 20 seconds.
-
Jan 20, 2023
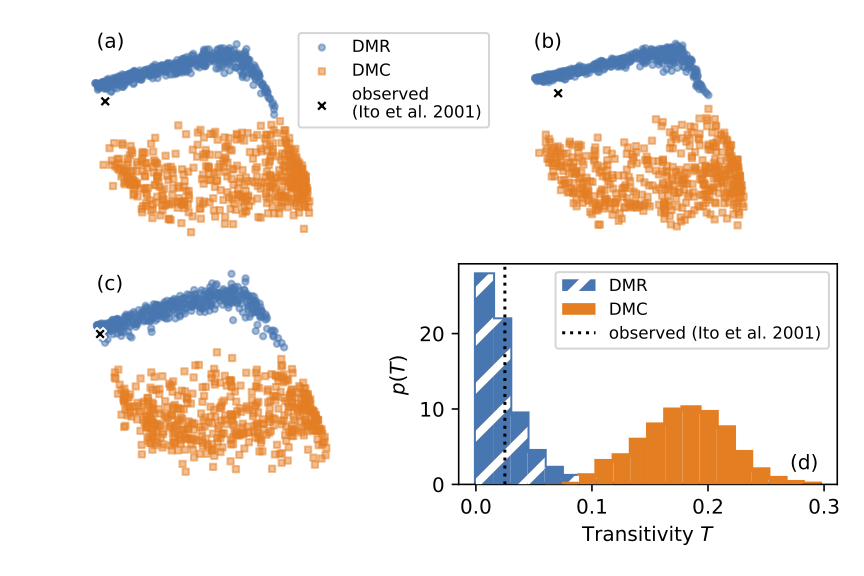 Selecting a small set of informative features from a large number of possibly noisy candidates is a challenging problem with many applications in machine learning and approximate Bayesian computation. In practice, the cost of computing informative features also needs to be considered. This is particularly important for networks because the computational costs of individual features can span several orders of magnitude. We addressed this issue for the network model selection problem using two approaches. First, we adapted nine feature selection methods to account for the cost of features. We show for two classes of network models that the cost can be reduced by two orders of magnitude without considerably affecting classification accuracy (proportion of correctly identified models). Second, we selected features using pilot simulations with smaller networks. This approach reduced the computational cost by a factor of 50 without affecting classification accuracy. To demonstrate the utility of our approach, we applied it to three different yeast protein interaction networks and identified the best-fitting duplication divergence model. Supplementary materials, including computer code to reproduce our results, are available online.
Selecting a small set of informative features from a large number of possibly noisy candidates is a challenging problem with many applications in machine learning and approximate Bayesian computation. In practice, the cost of computing informative features also needs to be considered. This is particularly important for networks because the computational costs of individual features can span several orders of magnitude. We addressed this issue for the network model selection problem using two approaches. First, we adapted nine feature selection methods to account for the cost of features. We show for two classes of network models that the cost can be reduced by two orders of magnitude without considerably affecting classification accuracy (proportion of correctly identified models). Second, we selected features using pilot simulations with smaller networks. This approach reduced the computational cost by a factor of 50 without affecting classification accuracy. To demonstrate the utility of our approach, we applied it to three different yeast protein interaction networks and identified the best-fitting duplication divergence model. Supplementary materials, including computer code to reproduce our results, are available online.
-
Jul 25, 2022
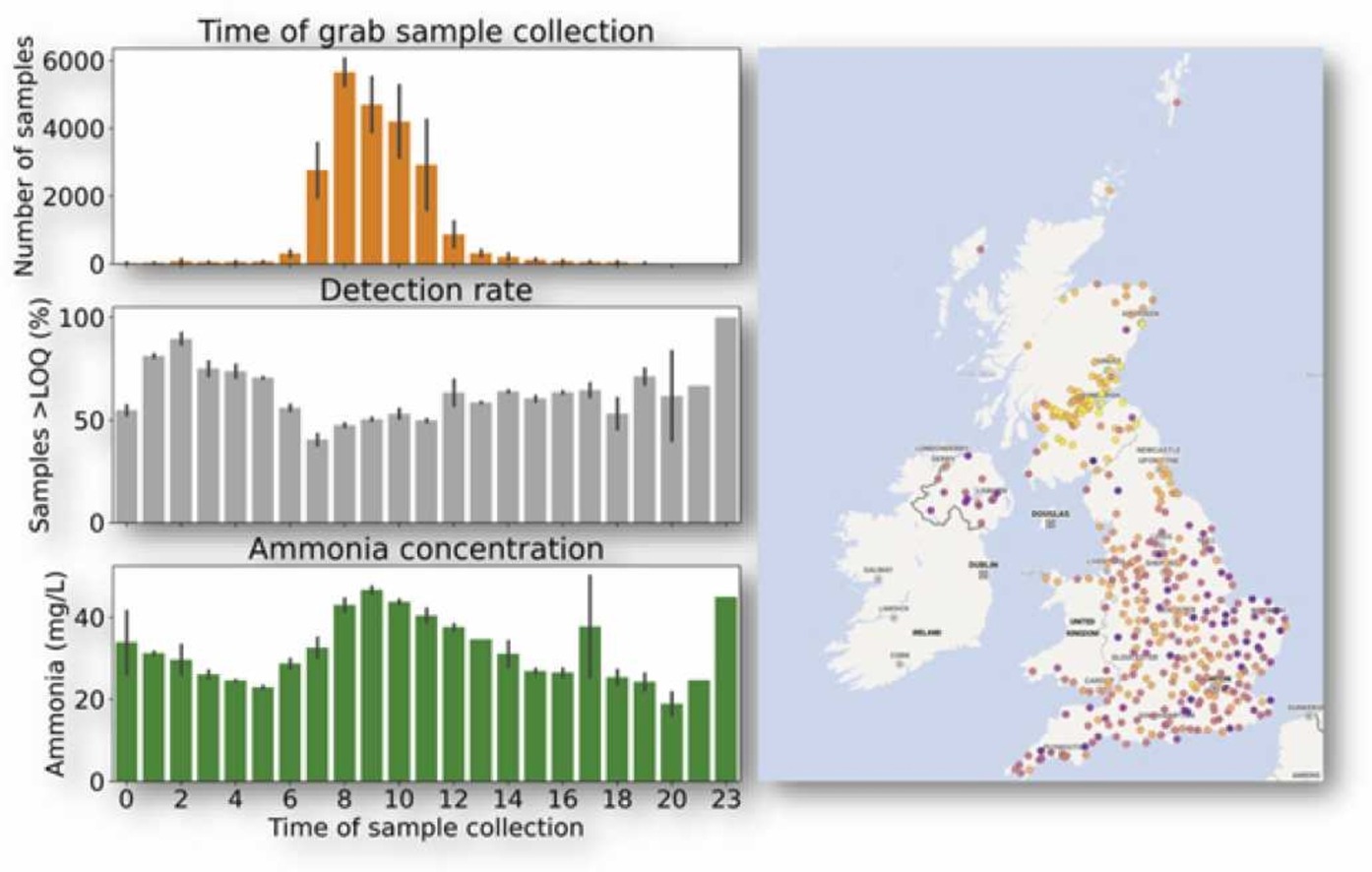 Accurate surveillance of the COVID-19 pandemic can be weakened by under-reporting of cases, particularly due to asymptomatic or pre-symptomatic infections, resulting in bias. Quantification of SARS-CoV-2 RNA in wastewater can be used to infer infection prevalence, but uncertainty in sensitivity and considerable variability has meant that accurate measurement remains elusive. Here, we use data from 45 sewage sites in England, covering 31% of the population, and estimate SARS-CoV-2 prevalence to within 1.1% of estimates from representative prevalence surveys (with 95% confidence). Using machine learning and phenomenological models, we show that differences between sampled sites, particularly the wastewater flow rate, influence prevalence estimation and require careful interpretation. We find that SARS-CoV-2 signals in wastewater appear 4–5 days earlier in comparison to clinical testing data but are coincident with prevalence surveys suggesting that wastewater surveillance can be a leading indicator for symptomatic viral infections. Surveillance for viruses in wastewater complements and strengthens clinical surveillance, with significant implications for public health.
Accurate surveillance of the COVID-19 pandemic can be weakened by under-reporting of cases, particularly due to asymptomatic or pre-symptomatic infections, resulting in bias. Quantification of SARS-CoV-2 RNA in wastewater can be used to infer infection prevalence, but uncertainty in sensitivity and considerable variability has meant that accurate measurement remains elusive. Here, we use data from 45 sewage sites in England, covering 31% of the population, and estimate SARS-CoV-2 prevalence to within 1.1% of estimates from representative prevalence surveys (with 95% confidence). Using machine learning and phenomenological models, we show that differences between sampled sites, particularly the wastewater flow rate, influence prevalence estimation and require careful interpretation. We find that SARS-CoV-2 signals in wastewater appear 4–5 days earlier in comparison to clinical testing data but are coincident with prevalence surveys suggesting that wastewater surveillance can be a leading indicator for symptomatic viral infections. Surveillance for viruses in wastewater complements and strengthens clinical surveillance, with significant implications for public health.
-
Jun 6, 2022
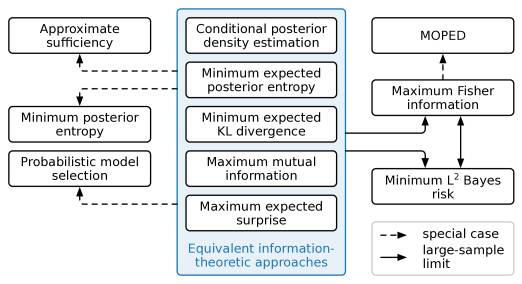 Extracting low-dimensional summary statistics from large datasets is essential for efficient (likelihood-free) inference. We propose obtaining summary statistics by minimizing the expected posterior entropy (EPE) under the prior predictive distribution of the model. We show that minimizing the EPE is equivalent to learning a conditional density estimator for the posterior as well as other information-theoretic approaches. Further summary extraction methods (including minimizing the $L^2$ Bayes risk, maximizing the Fisher information, and model selection approaches) are special or limiting cases of EPE minimization. We demonstrate that the approach yields high fidelity summary statistics by applying it to both a synthetic benchmark as well as a population genetics problem. We not only offer concrete recommendations for practitioners but also provide a unifying perspective for obtaining informative summary statistics.
Extracting low-dimensional summary statistics from large datasets is essential for efficient (likelihood-free) inference. We propose obtaining summary statistics by minimizing the expected posterior entropy (EPE) under the prior predictive distribution of the model. We show that minimizing the EPE is equivalent to learning a conditional density estimator for the posterior as well as other information-theoretic approaches. Further summary extraction methods (including minimizing the $L^2$ Bayes risk, maximizing the Fisher information, and model selection approaches) are special or limiting cases of EPE minimization. We demonstrate that the approach yields high fidelity summary statistics by applying it to both a synthetic benchmark as well as a population genetics problem. We not only offer concrete recommendations for practitioners but also provide a unifying perspective for obtaining informative summary statistics.
 Large-scale network data can pose computational challenges, be expensive to acquire, and compromise the privacy of individuals in social networks. We show that the locations and scales of latent space cluster models can be inferred from the number of connections between groups alone. We demonstrate this modelling approach using synthetic data and apply it to friendships between students collected as part of the Add Health study, eliminating the need for node-level connection data. The method thus protects the privacy of individuals and simplifies data sharing. It also offers performance advantages over node-level latent space models because the computational cost scales with the number of clusters rather than the number of nodes.
Large-scale network data can pose computational challenges, be expensive to acquire, and compromise the privacy of individuals in social networks. We show that the locations and scales of latent space cluster models can be inferred from the number of connections between groups alone. We demonstrate this modelling approach using synthetic data and apply it to friendships between students collected as part of the Add Health study, eliminating the need for node-level connection data. The method thus protects the privacy of individuals and simplifies data sharing. It also offers performance advantages over node-level latent space models because the computational cost scales with the number of clusters rather than the number of nodes. Gaussian processes (GPs) are sophisticated distributions to model functional data. Whilst theoretically appealing, they are computationally cumbersome except for small datasets. We implement two methods for scaling GP inference in Stan: First, a general sparse approximation using a directed acyclic dependency graph. Second, a fast, exact method for regularly spaced data modeled by GPs with stationary kernels using the fast Fourier transform. Based on benchmark experiments, we offer guidance for practitioners to decide between different methods and parameterizations. We consider two real-world examples to illustrate the package. The implementation follows Stan’s design and exposes performant inference through a familiar interface. Full posterior inference for more than ten thousand data points is feasible on a laptop in less than 20 seconds.
Gaussian processes (GPs) are sophisticated distributions to model functional data. Whilst theoretically appealing, they are computationally cumbersome except for small datasets. We implement two methods for scaling GP inference in Stan: First, a general sparse approximation using a directed acyclic dependency graph. Second, a fast, exact method for regularly spaced data modeled by GPs with stationary kernels using the fast Fourier transform. Based on benchmark experiments, we offer guidance for practitioners to decide between different methods and parameterizations. We consider two real-world examples to illustrate the package. The implementation follows Stan’s design and exposes performant inference through a familiar interface. Full posterior inference for more than ten thousand data points is feasible on a laptop in less than 20 seconds. Selecting a small set of informative features from a large number of possibly noisy candidates is a challenging problem with many applications in machine learning and approximate Bayesian computation. In practice, the cost of computing informative features also needs to be considered. This is particularly important for networks because the computational costs of individual features can span several orders of magnitude. We addressed this issue for the network model selection problem using two approaches. First, we adapted nine feature selection methods to account for the cost of features. We show for two classes of network models that the cost can be reduced by two orders of magnitude without considerably affecting classification accuracy (proportion of correctly identified models). Second, we selected features using pilot simulations with smaller networks. This approach reduced the computational cost by a factor of 50 without affecting classification accuracy. To demonstrate the utility of our approach, we applied it to three different yeast protein interaction networks and identified the best-fitting duplication divergence model. Supplementary materials, including computer code to reproduce our results, are available online.
Selecting a small set of informative features from a large number of possibly noisy candidates is a challenging problem with many applications in machine learning and approximate Bayesian computation. In practice, the cost of computing informative features also needs to be considered. This is particularly important for networks because the computational costs of individual features can span several orders of magnitude. We addressed this issue for the network model selection problem using two approaches. First, we adapted nine feature selection methods to account for the cost of features. We show for two classes of network models that the cost can be reduced by two orders of magnitude without considerably affecting classification accuracy (proportion of correctly identified models). Second, we selected features using pilot simulations with smaller networks. This approach reduced the computational cost by a factor of 50 without affecting classification accuracy. To demonstrate the utility of our approach, we applied it to three different yeast protein interaction networks and identified the best-fitting duplication divergence model. Supplementary materials, including computer code to reproduce our results, are available online. Accurate surveillance of the COVID-19 pandemic can be weakened by under-reporting of cases, particularly due to asymptomatic or pre-symptomatic infections, resulting in bias. Quantification of SARS-CoV-2 RNA in wastewater can be used to infer infection prevalence, but uncertainty in sensitivity and considerable variability has meant that accurate measurement remains elusive. Here, we use data from 45 sewage sites in England, covering 31% of the population, and estimate SARS-CoV-2 prevalence to within 1.1% of estimates from representative prevalence surveys (with 95% confidence). Using machine learning and phenomenological models, we show that differences between sampled sites, particularly the wastewater flow rate, influence prevalence estimation and require careful interpretation. We find that SARS-CoV-2 signals in wastewater appear 4–5 days earlier in comparison to clinical testing data but are coincident with prevalence surveys suggesting that wastewater surveillance can be a leading indicator for symptomatic viral infections. Surveillance for viruses in wastewater complements and strengthens clinical surveillance, with significant implications for public health.
Accurate surveillance of the COVID-19 pandemic can be weakened by under-reporting of cases, particularly due to asymptomatic or pre-symptomatic infections, resulting in bias. Quantification of SARS-CoV-2 RNA in wastewater can be used to infer infection prevalence, but uncertainty in sensitivity and considerable variability has meant that accurate measurement remains elusive. Here, we use data from 45 sewage sites in England, covering 31% of the population, and estimate SARS-CoV-2 prevalence to within 1.1% of estimates from representative prevalence surveys (with 95% confidence). Using machine learning and phenomenological models, we show that differences between sampled sites, particularly the wastewater flow rate, influence prevalence estimation and require careful interpretation. We find that SARS-CoV-2 signals in wastewater appear 4–5 days earlier in comparison to clinical testing data but are coincident with prevalence surveys suggesting that wastewater surveillance can be a leading indicator for symptomatic viral infections. Surveillance for viruses in wastewater complements and strengthens clinical surveillance, with significant implications for public health. Extracting low-dimensional summary statistics from large datasets is essential for efficient (likelihood-free) inference. We propose obtaining summary statistics by minimizing the expected posterior entropy (EPE) under the prior predictive distribution of the model. We show that minimizing the EPE is equivalent to learning a conditional density estimator for the posterior as well as other information-theoretic approaches. Further summary extraction methods (including minimizing the $L^2$ Bayes risk, maximizing the Fisher information, and model selection approaches) are special or limiting cases of EPE minimization. We demonstrate that the approach yields high fidelity summary statistics by applying it to both a synthetic benchmark as well as a population genetics problem. We not only offer concrete recommendations for practitioners but also provide a unifying perspective for obtaining informative summary statistics.
Extracting low-dimensional summary statistics from large datasets is essential for efficient (likelihood-free) inference. We propose obtaining summary statistics by minimizing the expected posterior entropy (EPE) under the prior predictive distribution of the model. We show that minimizing the EPE is equivalent to learning a conditional density estimator for the posterior as well as other information-theoretic approaches. Further summary extraction methods (including minimizing the $L^2$ Bayes risk, maximizing the Fisher information, and model selection approaches) are special or limiting cases of EPE minimization. We demonstrate that the approach yields high fidelity summary statistics by applying it to both a synthetic benchmark as well as a population genetics problem. We not only offer concrete recommendations for practitioners but also provide a unifying perspective for obtaining informative summary statistics.